Chapter 5 layouts
5.1 choosing a layout
Depending on what you want to show, different layouts help you in achieving that goal. The list of layouts in igraph is:
ggpraph(data, layout = "[insert name algorithm]") ...
| name | explanation | when |
|---|---|---|
| bipartite | [Simple two-row layout for bipartite graphs] | two independent groups |
| circle | [circle shaped] | equal importance of nodes, equal distances facilitate communication, small networks |
| dh | [Davidson-Harel layout algorithm] | force-directed layout that lead to closely connected nodes being closer to one another, loosely connected nodes pushed towards outside |
| drl | [DrL graph layout generator] | force-directed layout for large graphs that lead to closely connected nodes being closer to one another, loosely connected nodes pushed towards outside |
| fr | [Fruchterman-Reingold layout algorithm] | force-directed layout that lead to closely connected nodes being closer to one another, loosely connected nodes pushed towards outside |
| grid | [simple grid layout] | small networks, fixed layout easy to interpret |
| gem | [GEM layout algorithm] | force-directed layout that lead to closely connected nodes being closer to one another, loosely connected nodes pushed towards outside |
| graphopt | [graphopt layout algorithm] | force-directed layout for large graphs that lead to closely connected nodes being closer to one another, loosely connected nodes pushed towards outside |
| kk | [Kamada-Kawai layout algorithm] | force-directed layout that lead to closely connected nodes being closer to one another, loosely connected nodes pushed towards outside |
| lgl | [Large Graph Layout] | force-directed layout for very large graphs that lead to closely connected nodes being closer to one another, loosely connected nodes pushed towards outside |
| mds | [layout by multidimensional scaling] | when you want to use multiple measures of distance between nodes to determine layout |
| nicely | [picks one for you based on data] | useful start |
| randomly | [random placement] | never |
| sugiyama | [Sugiyama graph layout generator] | layered/hierarchical network where there is an ordered sequence of events/actions/relationships. Designed for layered directed acyclic graphs, and minimises edge crossings |
| sphere | [sphere shaped] | not sure, small networks |
| star | [star shaped] | not sure, small networks |
| tree | [Reingold-Tilford graph layout algorithm] | hierarchical network (tree!) |
Let’s load in some data.
df_edges <- data.frame(
from = c("Gert","Gert","Gert","Gert","Gert","Ben","Anne"),
to = c("Anne","Ben","Winy","Vera","Laura","Winy","Vera"),
closeness = c("very close", "close", "close", "close", "somewhat close",
"very close", "very close")
)
df_nodes <- data.frame(
name = c("Gert", "Anne", "Ben", "Winy", "Vera", "Laura"),
relation = c("Ego", "Partner", "Family", "Family", "Friend", "Friend")
)
df_network <- tbl_graph(nodes = df_nodes, edges = df_edges, directed = FALSE)It’s a small network, let’s try a circle:
ggraph(df_network, layout = "circle") +
geom_edge_link(aes(linetype = closeness)) +
geom_node_point(aes(colour = relation), size = 13) +
geom_node_text(aes(label = name), colour = "white") +
theme_void() +
scale_color_brewer(palette = "Set2")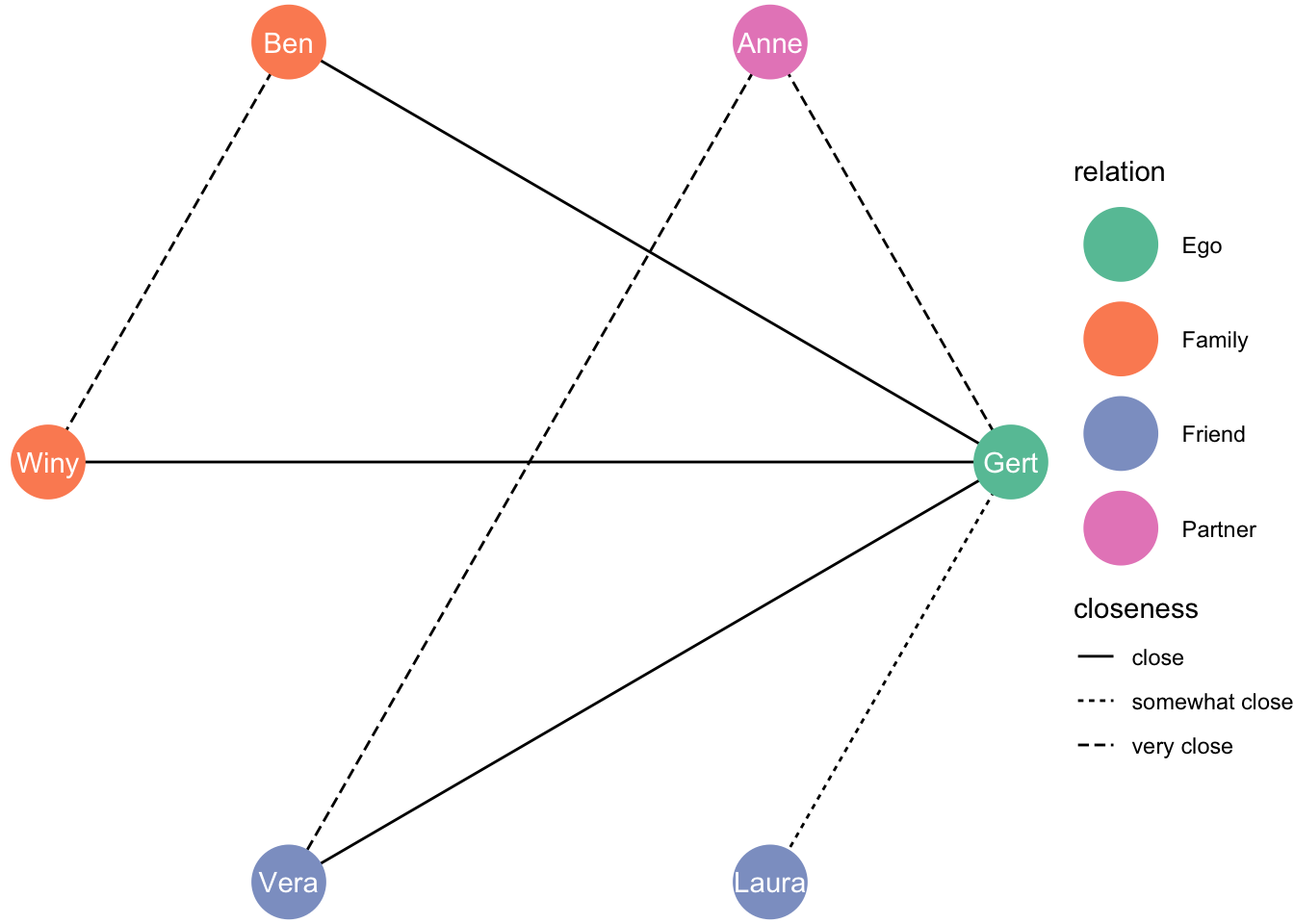
5.1.0.1 Circle with ego in the middle!
[this layout comes from the package graphlayouts]
ggraph(df_network, layout = "focus", focus = 1) +
geom_edge_link(aes(linetype = closeness)) +
geom_node_point(aes(colour = relation), size = 13) +
geom_node_text(aes(label = name), colour = "white") +
theme_void() +
scale_color_brewer(palette = "Set2")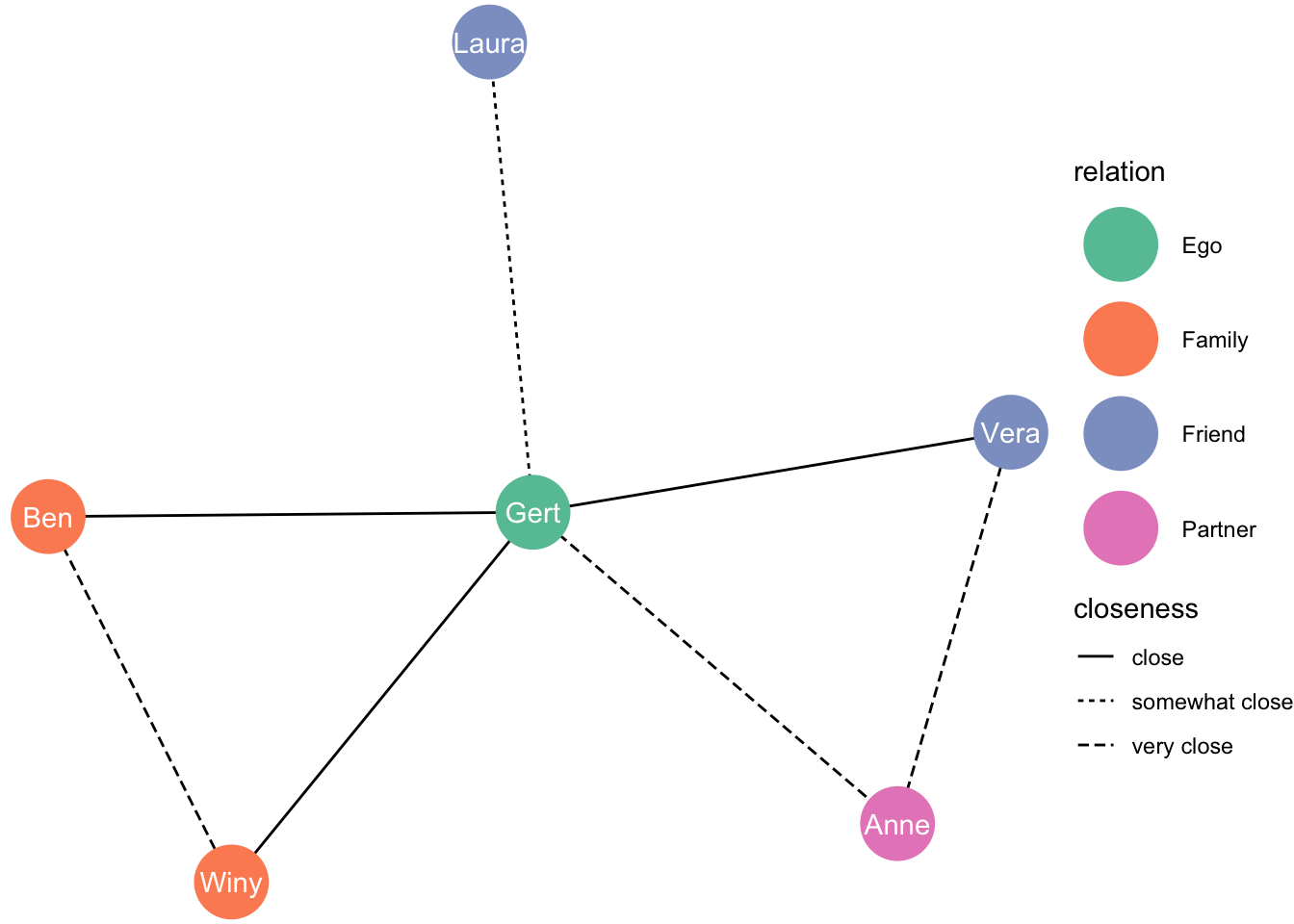
Or on a square?
ggraph(df_network, layout = "grid") +
geom_edge_link(aes(linetype = closeness)) +
geom_node_point(aes(colour = relation), size = 13) +
geom_node_text(aes(label = name), colour = "white") +
theme_void() +
scale_color_brewer(palette = "Set2")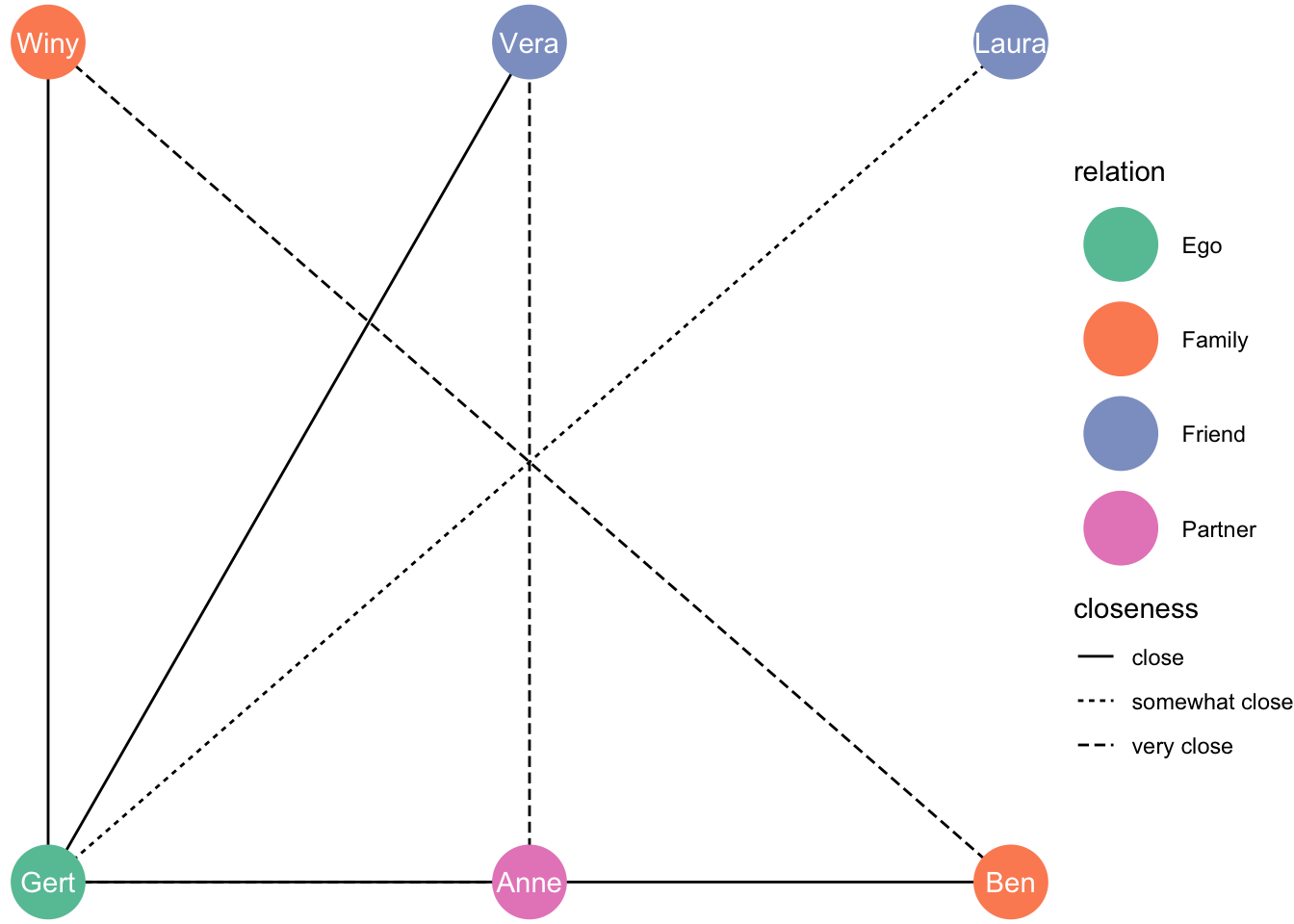
Using a force-directed algorithm (Fruchterman-Reingold layout algorithm) :
set.seed(1)
ggraph(df_network, layout = "fr") +
geom_edge_link(aes(linetype = closeness)) +
geom_node_point(aes(colour = relation), size = 13) +
geom_node_text(aes(label = name), colour = "white") +
theme_void() +
scale_color_brewer(palette = "Set2")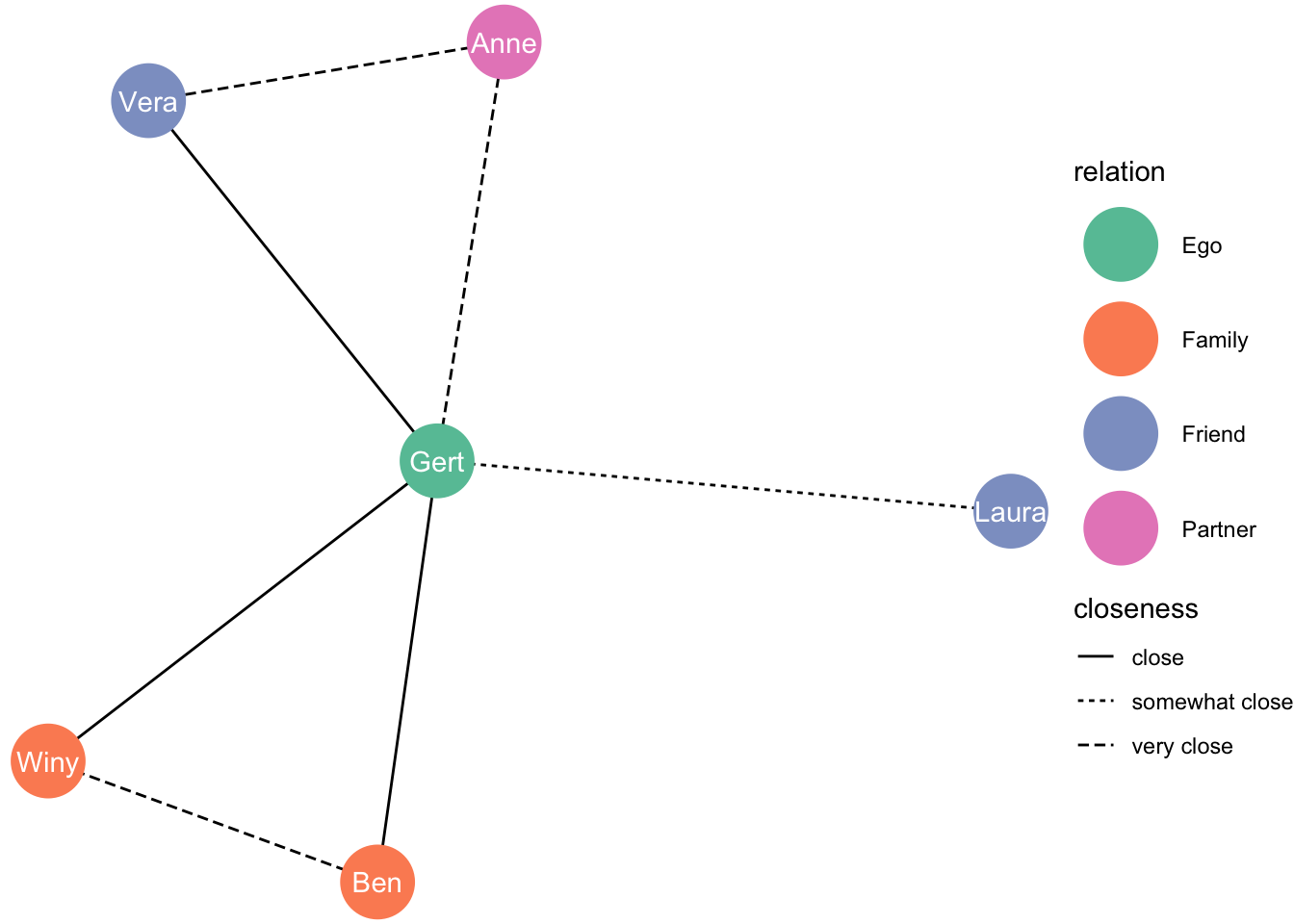
Slightly different each time:
set.seed(2)
ggraph(df_network, layout = "fr") +
geom_edge_link(aes(linetype = closeness)) +
geom_node_point(aes(colour = relation), size = 13) +
geom_node_text(aes(label = name), colour = "white") +
theme_void() +
scale_color_brewer(palette = "Set2")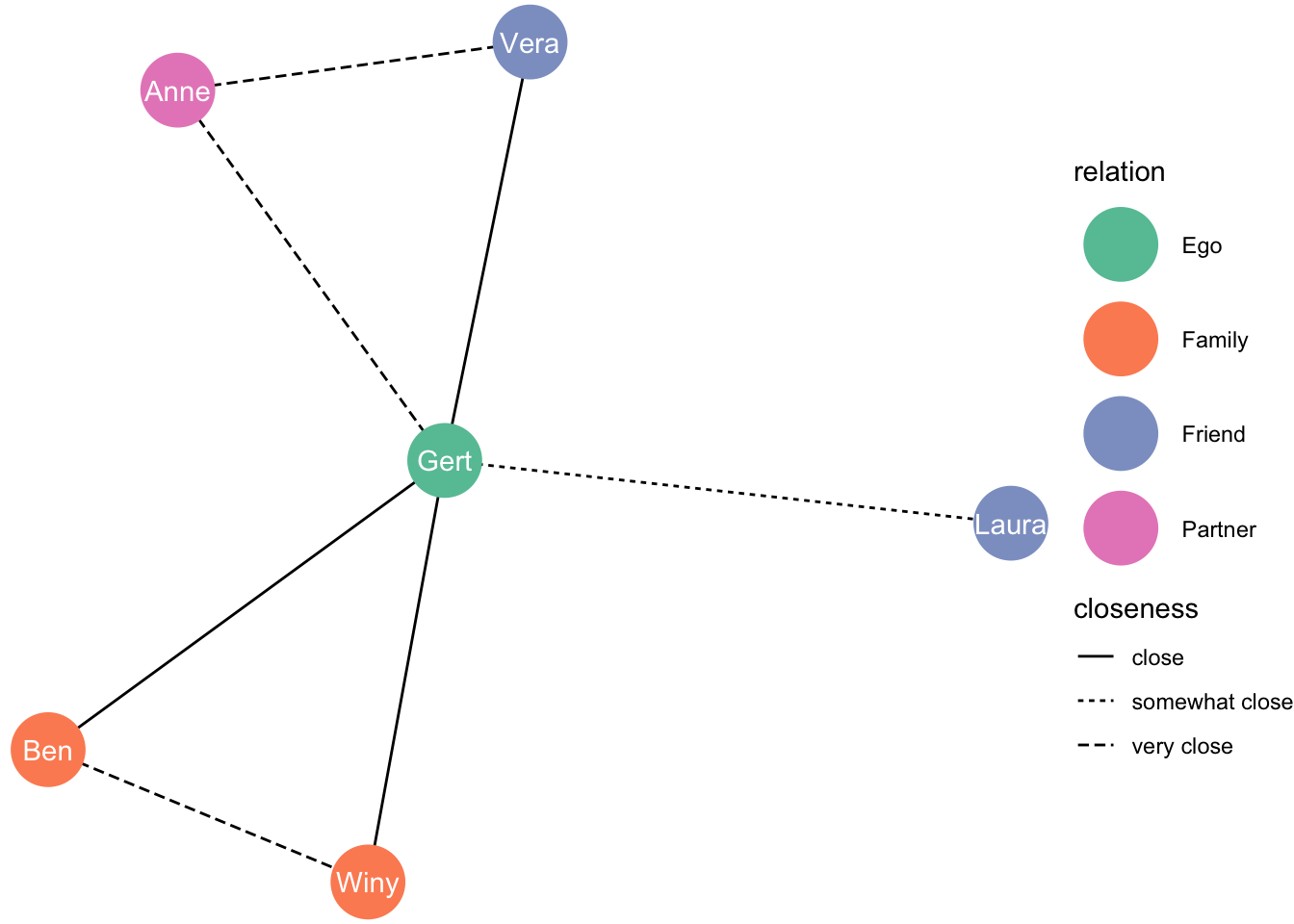
Let’s look at larger graphs on … dolphins! 190 of them. First the data.
library(igraph)
dolphins_graphml <- igraph::read.graph("https://raw.githubusercontent.com/bansallab/asnr/master/Networks/Mammalia/dolphin_association_weighted/weighted_FORAGE_dolphin_florida.graphml", format = "graphml")
dolphins <- as_tbl_graph(dolphins_graphml)Let’s try putting them in a circle.
ggraph(dolphins, layout = "circle") +
geom_edge_link() +
geom_node_point() +
theme_graph() +
coord_fixed()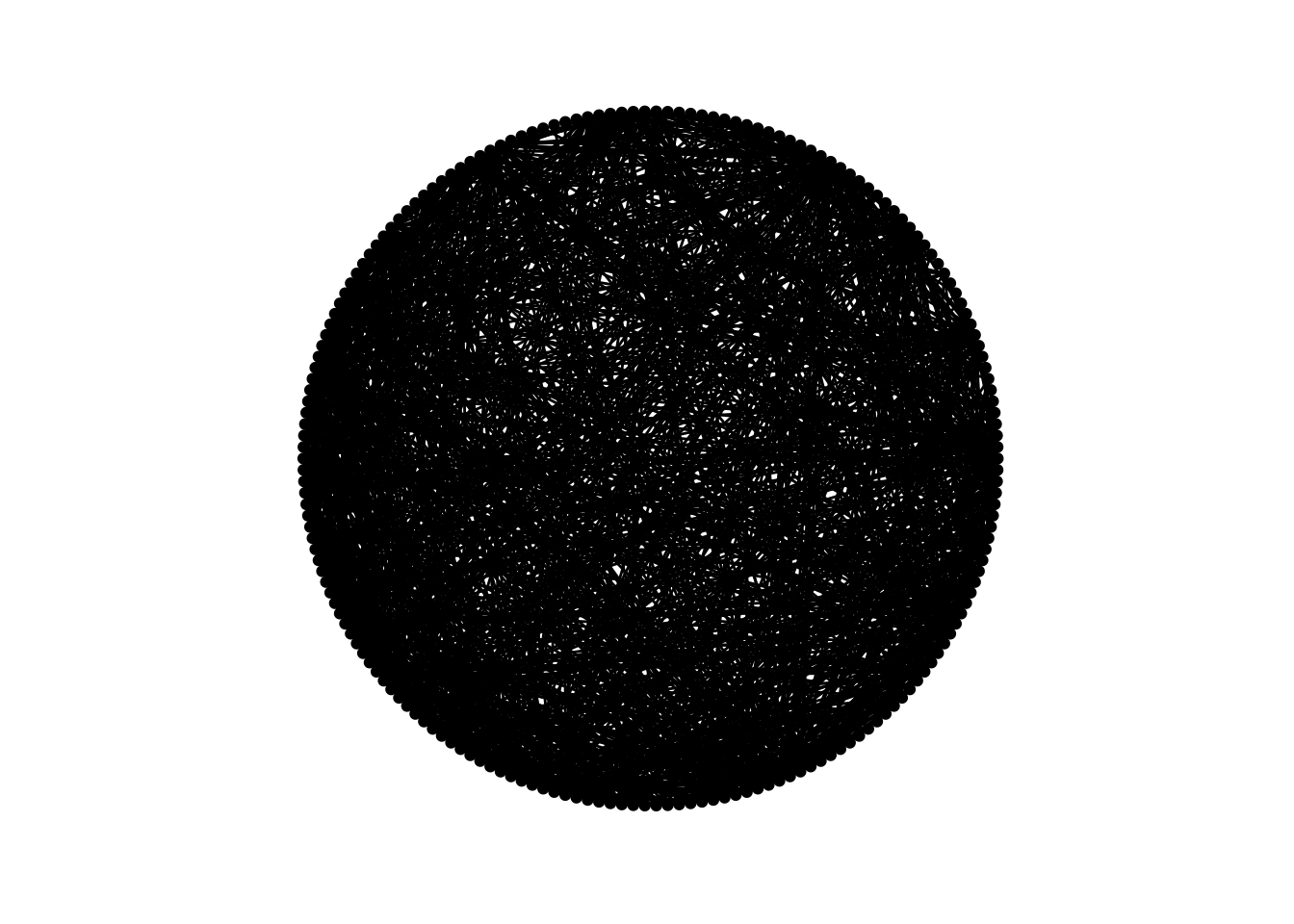
Ok…
ggraph(dolphins, layout = "circle") +
geom_edge_link(alpha = 0.1) +
geom_node_point() +
theme_graph() +
coord_fixed()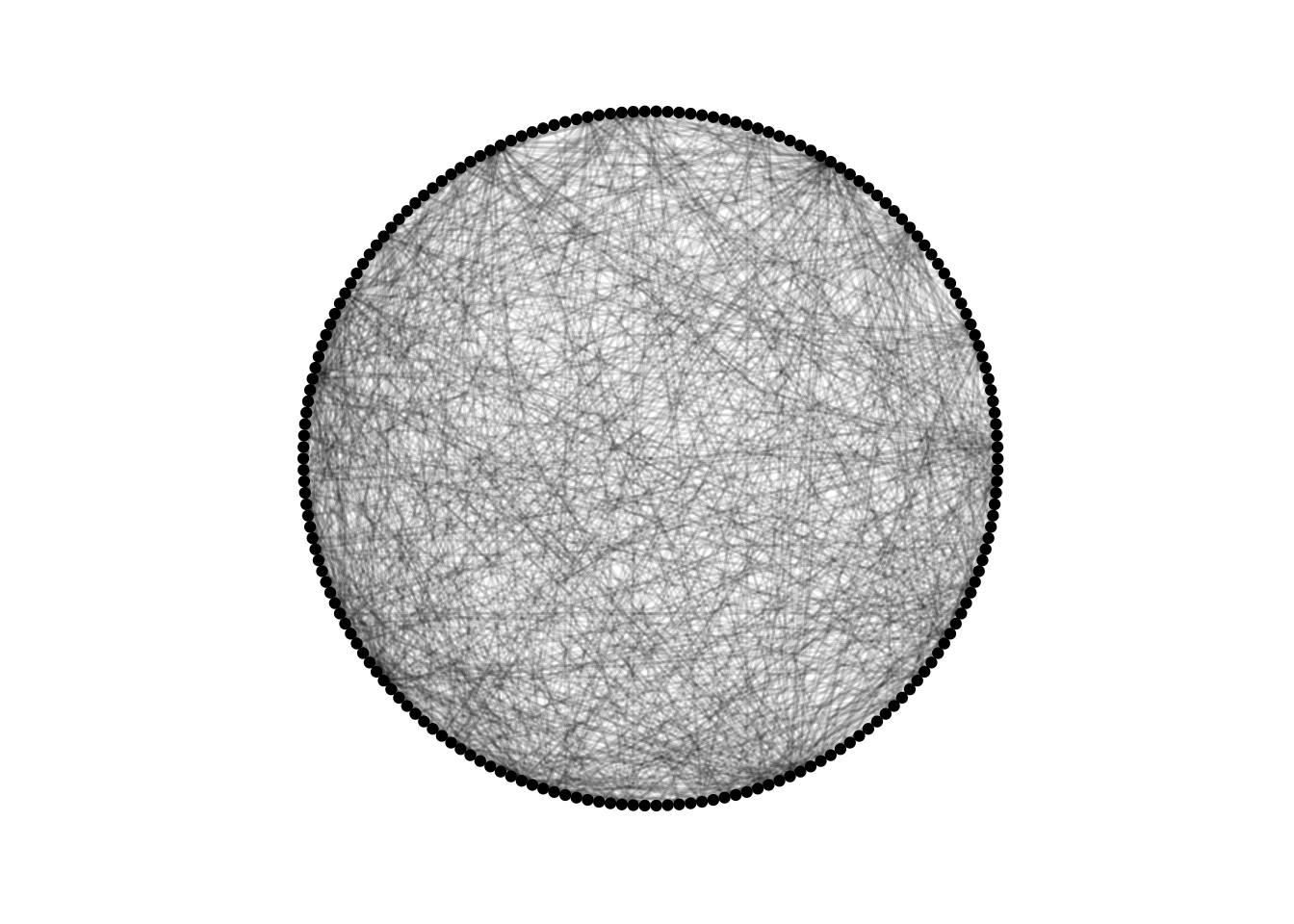
Pretty but useless!
Let’s try these force-directed layouts that handle large networks better.
ggraph(dolphins, layout = "kk") +
geom_edge_link() +
geom_node_point() +
theme_graph() 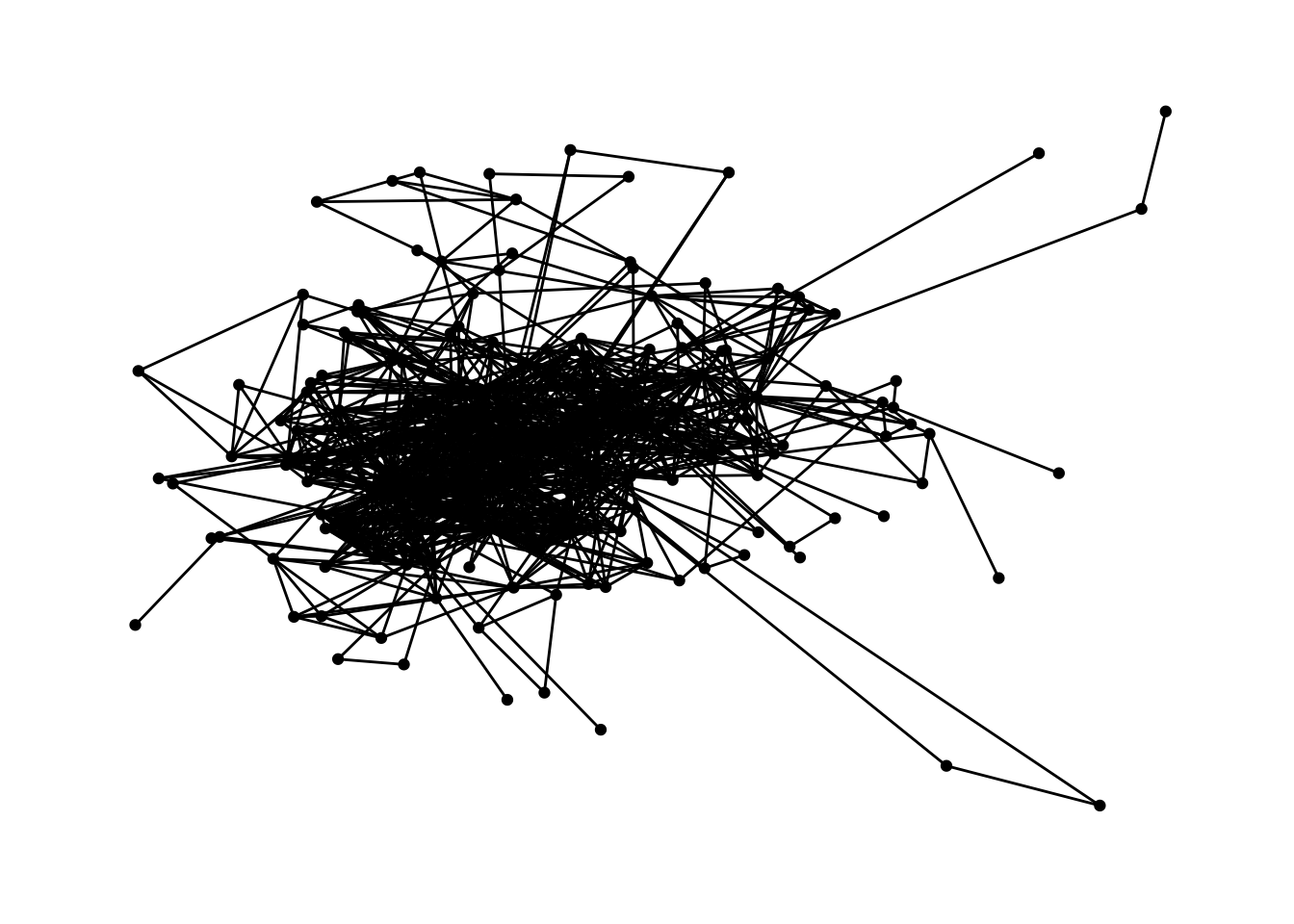
The many edges make these graphs problematic. Some layout alghorithms reduced the number of edges/ties. We opt for making them very light in colour, and see through:
ggraph(dolphins, layout = "kk") +
geom_edge_link(alpha = 0.1) +
geom_node_point() +
theme_graph() 
ggraph(dolphins, layout = "fr") +
geom_edge_link(alpha = 0.1) +
geom_node_point() +
theme_graph() 
ggraph(dolphins, layout = "dh") +
geom_edge_link(alpha = 0.1) +
geom_node_point() +
theme_graph() 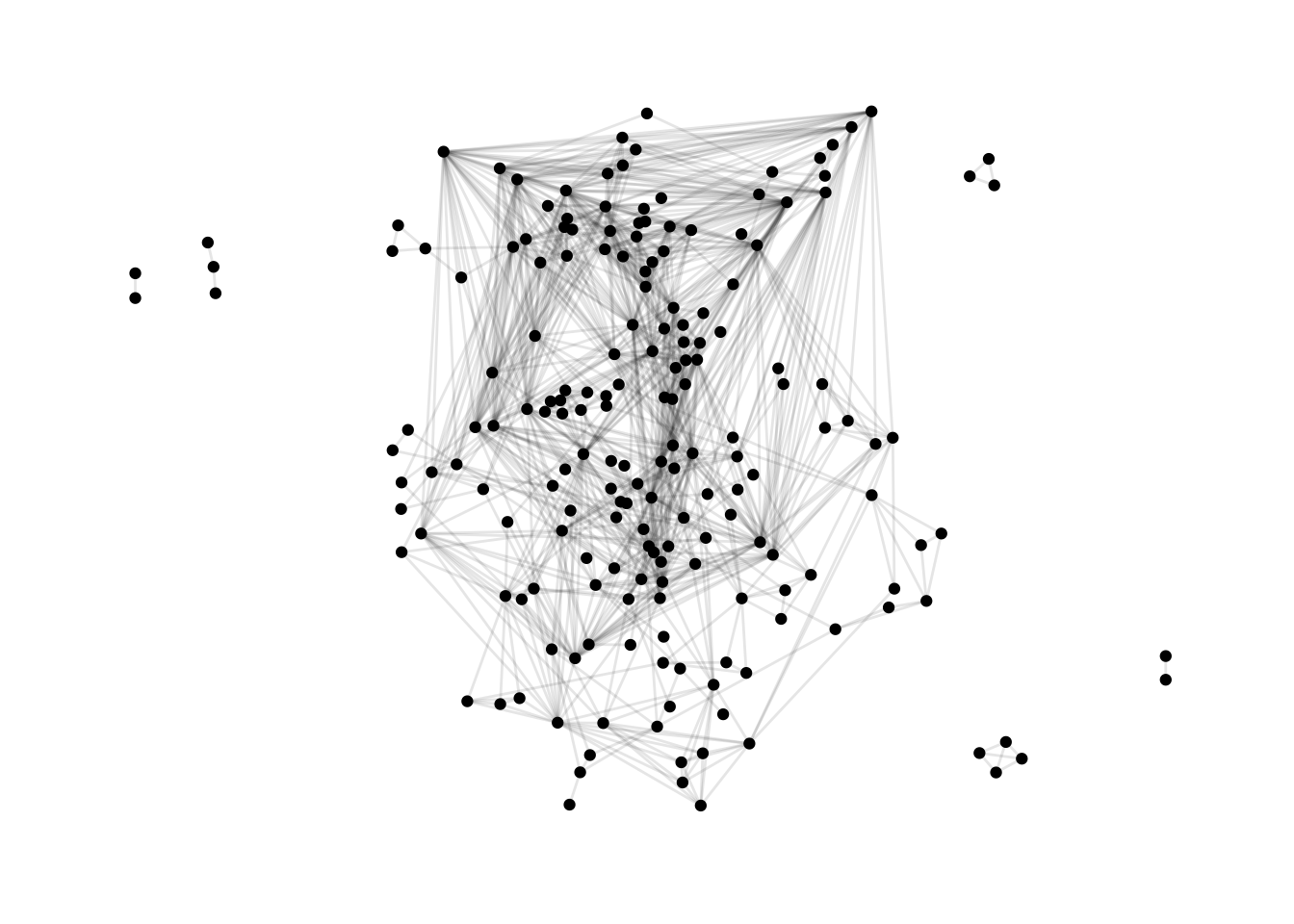
5.1.1 Keeping a layout for future use
Remember this graph?
set.seed(1)
ggraph(df_network, layout = "fr") +
geom_edge_link(aes(linetype = closeness)) +
geom_node_point(aes(colour = relation), size = 13) +
geom_node_text(aes(label = name), colour = "white") +
theme_void() +
scale_color_brewer(palette = "Set2")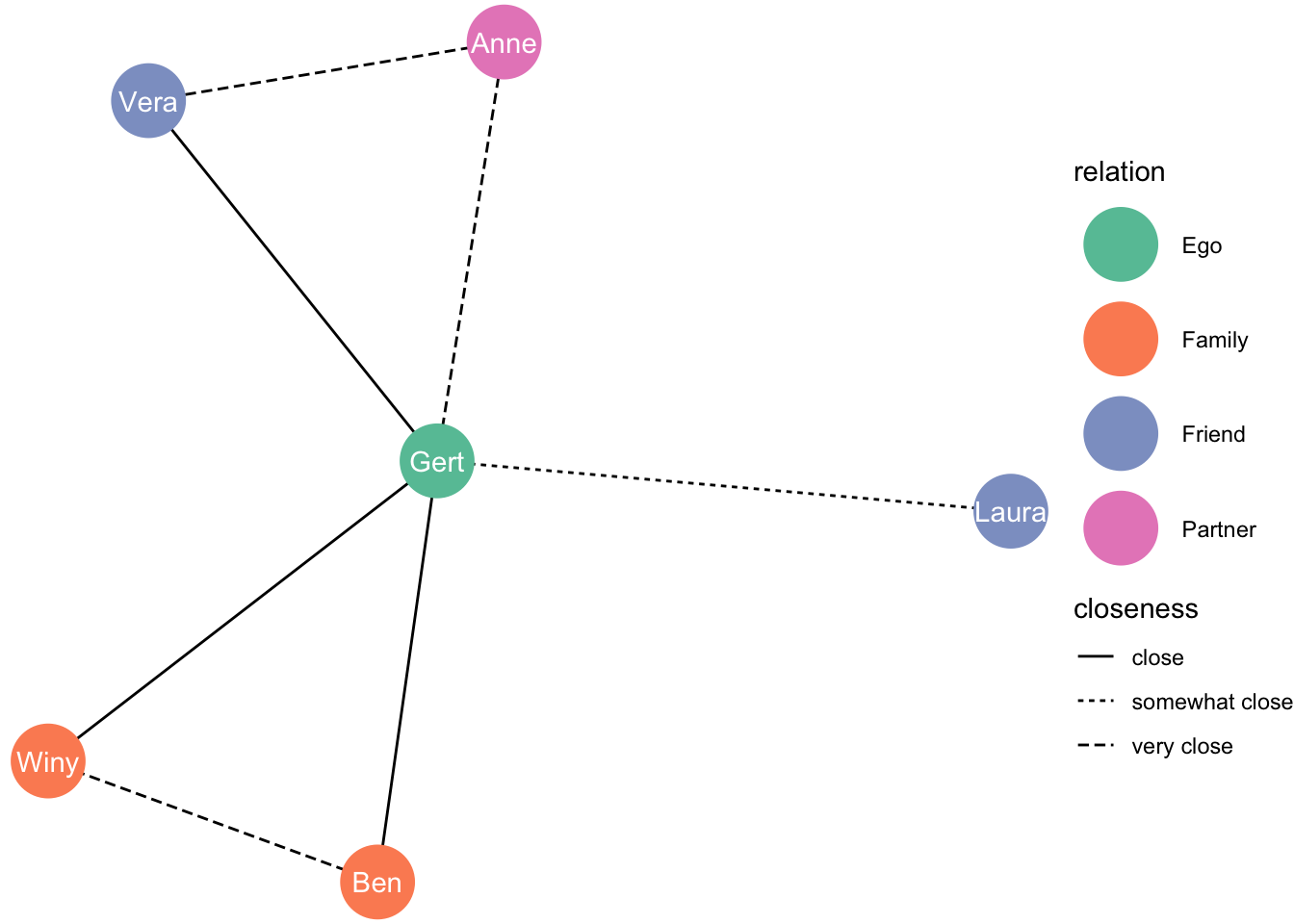
Let’s say I have also information on the frequency of contact, and I also would like to show this information in a graph, then it would be useful to have exactly the same layout. Let’s see how to do this:
First, let’s create the new information on frequency of contact:
df_network <- df_network %>%
activate(edges) %>%
mutate( # Create new variable in edges dataset
freq_cont = c("daily", "monthly", "monthly", "monthly", "weekly",
"daily", "monthly")
)We’ve indeed managed to create new variable:
df_network## # A tbl_graph: 6 nodes and 7 edges
## #
## # An undirected simple graph with 1 component
## #
## # Edge Data: 7 × 4 (active)
## from to closeness freq_cont
## <int> <int> <chr> <chr>
## 1 1 2 very close daily
## 2 1 3 close monthly
## 3 1 4 close monthly
## 4 1 5 close monthly
## 5 1 6 somewhat close weekly
## 6 3 4 very close daily
## # … with 1 more row
## #
## # Node Data: 6 × 2
## name relation
## <chr> <chr>
## 1 Gert Ego
## 2 Anne Partner
## 3 Ben Family
## # … with 3 more rowsNow, we want two graphs with the same layout, one for closeness, and one for frequency of contact. We need to keep the layout that the algorithm that determined our layout is kept:
set.seed(1)
layout_nw <- create_layout(df_network, layout = 'fr')Just a simple dataframe:
layout_nw## x y name relation .ggraph.orig_index circular .ggraph.index
## 1 7.689453 11.83531 Gert Ego 1 FALSE 1
## 2 7.858226 13.19301 Anne Partner 2 FALSE 2
## 3 7.539544 10.47034 Ben Family 3 FALSE 3
## 4 6.709893 10.86258 Winy Family 4 FALSE 4
## 5 6.962720 13.00292 Vera Friend 5 FALSE 5
## 6 9.134448 11.67227 Laura Friend 6 FALSE 6Let’s graph:
closeness_graph <- ggraph(layout_nw) +
geom_edge_link(aes(linetype = closeness)) +
geom_node_point(aes(colour = relation), size = 13) +
geom_node_text(aes(label = name), colour = "white") +
theme_void() +
scale_color_brewer(palette = "Set2")
closeness_graph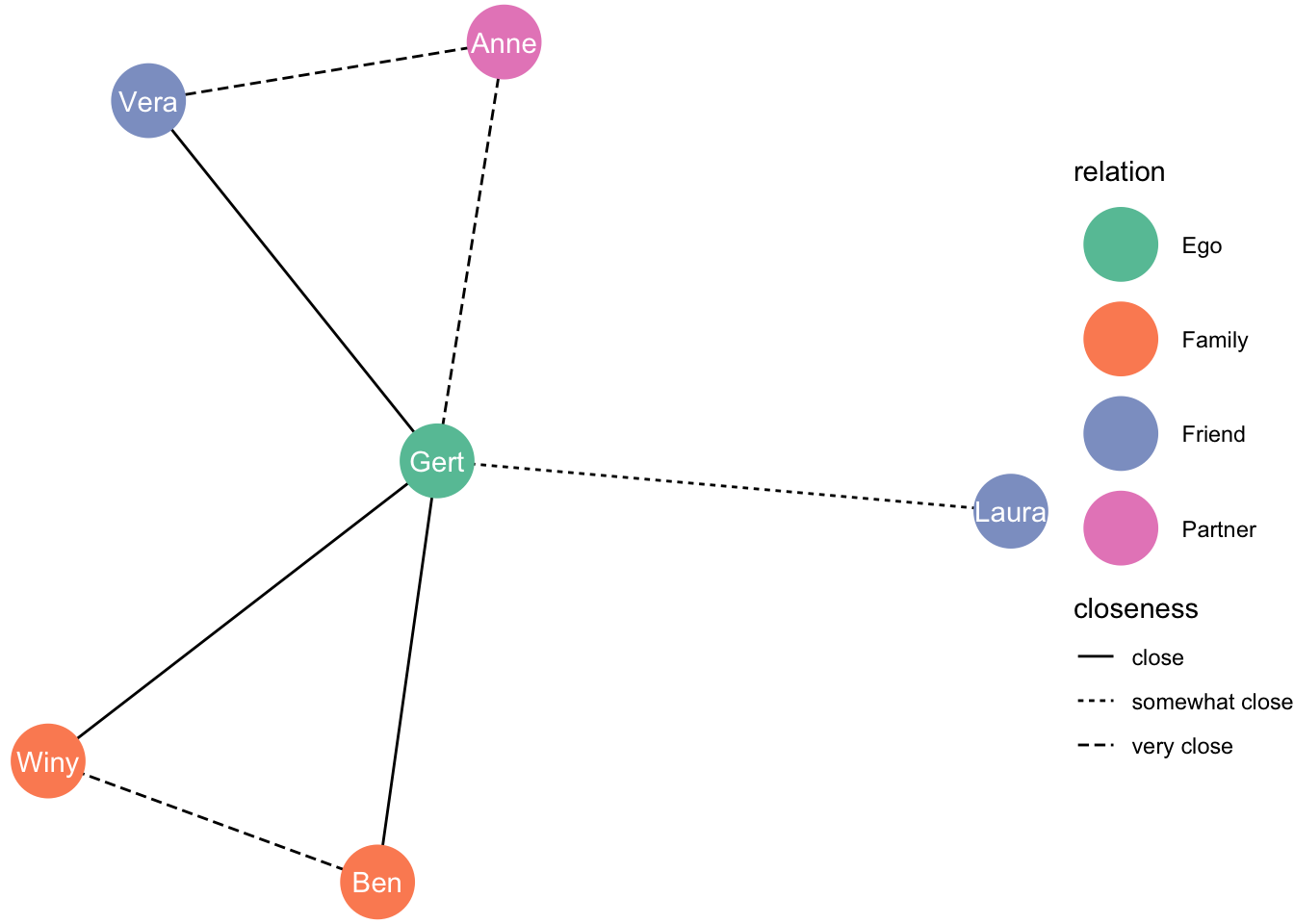
Now let’s graph frequency of contact
freq_cont_graph <- ggraph(layout_nw) +
geom_edge_link(aes(linetype = freq_cont)) +
geom_node_point(aes(colour = relation), size = 13) +
geom_node_text(aes(label = name), colour = "white") +
theme_void() +
scale_color_brewer(palette = "Set2")
freq_cont_graph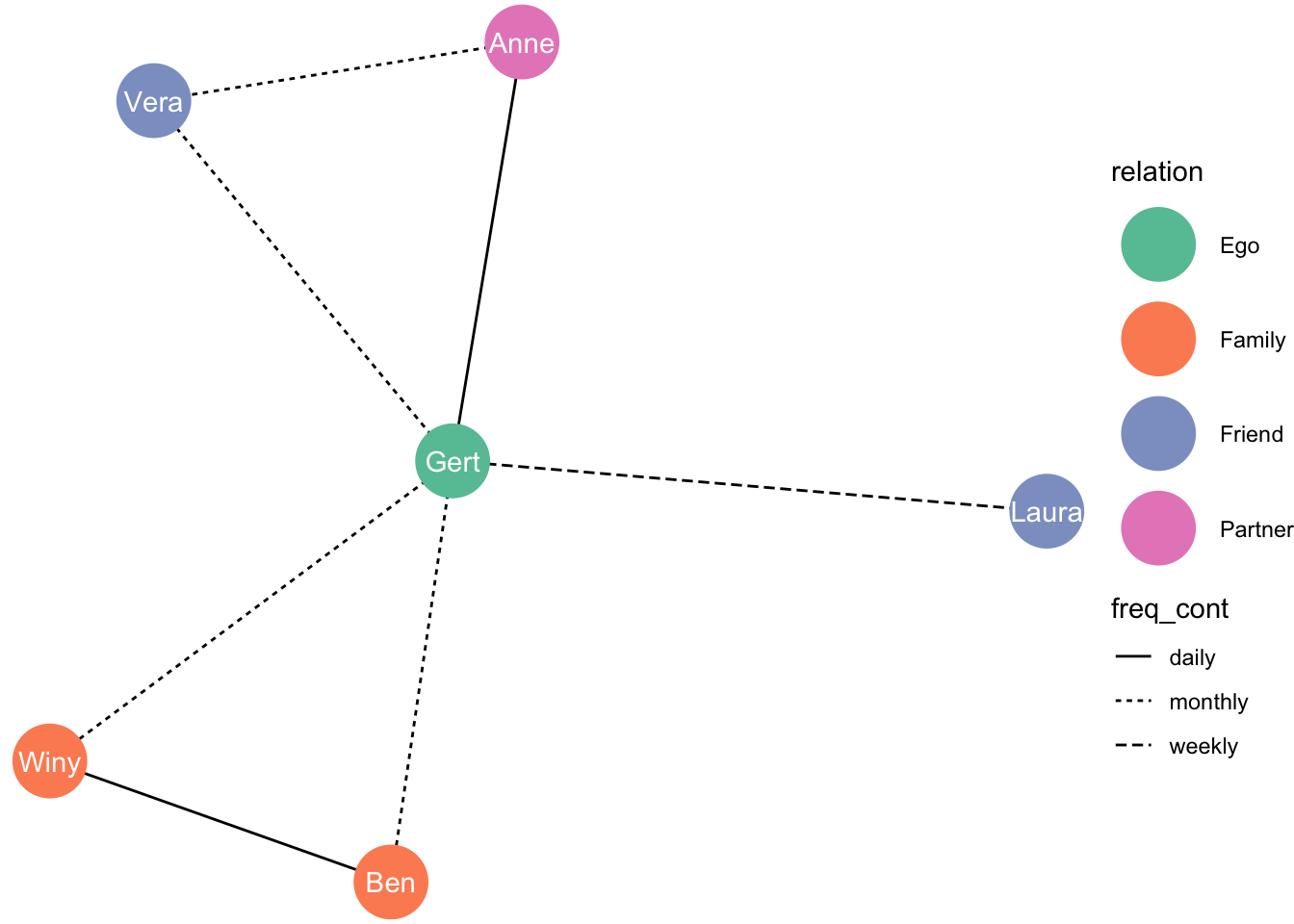
Let’s put them next to one another:
library(patchwork)
closeness_graph + freq_cont_graph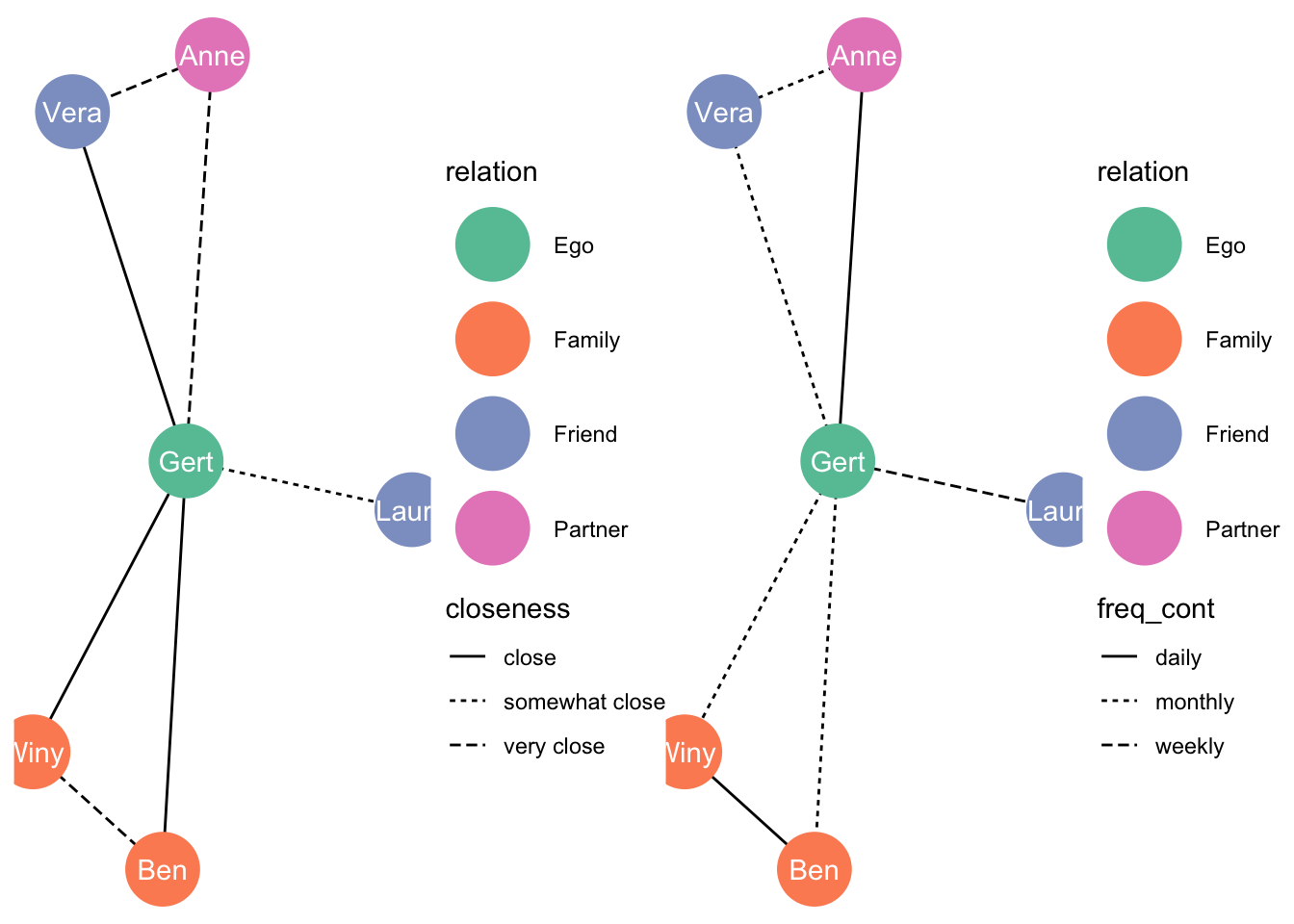
5.2 DIY
Here is data from Zebra!
zebra_graphml <- read.graph("https://raw.githubusercontent.com/bansallab/asnr/master/Networks/Mammalia/zebra_groupmembership_weighted/zebra_sundaresan_interaction_attribute.graphml", format = "graphml")
zebra <- as_tbl_graph(zebra_graphml)Try to find a useful layout